Abstract
INTRODUCTION : Multiple myeloma (MM) is a plasma cell dyscrasia that is characterized by the presence of both a clonal plasma cell population in the bone marrow and intact monoclonal immunoglobulin and/or free light chain (LC) proteins in the serum. It has been shown that MM may be an ominous precursor to light chain-associated (AL) amyloidosis, a devastating protein misfolding disorder characterized by the systemic deposition of extracellular amyloid fibrils composed of LC components. In the US, there are ~30,000 new cases of MM every year, and it is estimated that 10 - 15% of these patients will progress to develop amyloidosis.
Light chain amyloid may deposit in any organ or tissue, but the heart, liver, spleen, kidneys and nerves are most commonly involved. The accumulation of amyloid in organs, especially the heart and kidneys, results in architectural damage leading to progressive organ dysfunction and death. It has been documented that concomitant MM with amyloidosis results in a considerably poorer prognosis than MM alone.
Currently, there is no way of identifying those MM patients who will progress to develop LC-associated amyloidosis, but the ability to identify at-risk patients would be of clinical value because anti-amyloid immunotherapeutic agents are on the horizon. If such patients are identified prior to the onset of amyloid-associated clinical symptoms and organ damage, physicians could implement closer monitoring and begin a maintenance dose of amyloid-clearing antibody reagents.
Early detection of amyloid is critical for enhancing patient survival; thus, we have investigated an approach to discern which MM patients have a greater likelihood of developing amyloidosis, based on the amyloid-related properties of the monoclonal light chain. We have previously shown that LC derived from MM and AL patients, when radiolabeled, can be recruited by synthetic amyloid-like fibrils. We observed that MM LC were recruited less effectively than AL LC, suggesting that there is disease specific reactivity of LC with amyloid fibrils (Martin et al . 2017 PLoS One, 12(3): e0174152.). We have since adapted this approach to a more facile and practical microplate assay measuring the ability of LC to compete with monomeric protein for fibril elongation. We have been able to discern patient populations with this technique.
MATERIALS & METHODS : For these studies, 96-well microplates were coated with 0.83 μM synthetic amyloid-like fibrils (rVλ6Wil) and dried at 37°C overnight. After washing and a "blocking" step, a 100 μL solution of urinary LC protein-either AL amyloidosis or MM patient-derived-at increasing serial concentrations (0.05 - 5 µg/mL) mixed with biotinylated rVλ6Wil protein monomer (5 nM) was added to the wells. After 1 h incubation, the amount of fibril-recruited biontinyl-rVλ6Wil monomer was measured. Concentration gradients for MM and AL LC were measured and compared using a one-tailed Student's t-test.
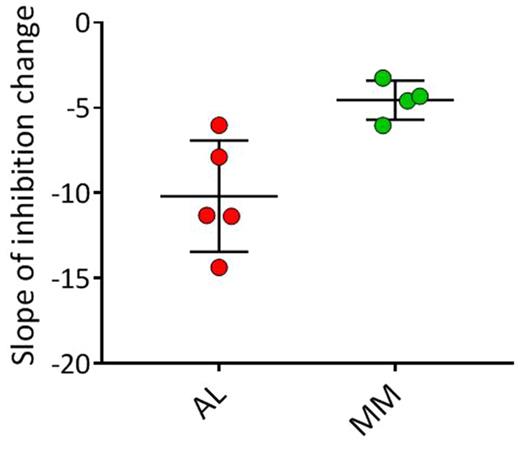
RESULTS : The data indicate that, in this competition assay, AL-associated urinary LC inhibited rVλ6Wil monomer recruitment more effectively than MM-derived LC. Our concentration-dependent gradient analysis discerned proteins from the two disease populations with statistical significance (p < 0.007). Notably, LC from a patient initially diagnosed with MM, who later developed amyloidosis, behaved as an AL LC in this assay, indicating the ability of this technique to predict the amyloidogenic propensity of LC.
DISCUSSION : We have studied the capacity of intact, patient-derived LC to inhibit synthetic fibril elongation as a means of predicting the risk of amyloidosis. The assay was able to discriminate between LC proteins derived from MM and AL patients. This approach, in conjunction with other metrics, may be useful in discerning those patients with MM, or perhaps LC-associated monoclonal gammopathy of undetermined significance (LCMGUS), who are at risk of developing amyloidosis based on LC activity in our recruitment assay. We are currently adapting this technique for use with patient serum.
Martin: University of Tennessee Research Foundation: Patents & Royalties. Wall: University of Tennessee Research Foundation: Patents & Royalties.
Author notes
Asterisk with author names denotes non-ASH members.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal